Kolekce Atom Index In Position_Restraints Out Of Bounds
Kolekce Atom Index In Position_Restraints Out Of Bounds. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. Atom index n in position_restraints out of bounds. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. A common problem is placing position restraint files for multiple molecules out of order.
Tady Team Stuttgart Model 2018 Igem Org
When i followed the manual, i encountered the following problems. file posre.itp, line 54 : file posre_protein_chain_h.itp, line 334 : Atom index (51) in position_restraints out of bounds. A common problem is placing position restraint files for multiple molecules out of order.Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order.
Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. file posre_protein_chain_h.itp, line 334 : Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. Atom index n in position_restraints out of bounds.
file posre_protein_chain_h.itp, line 334 : Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. When i followed the manual, i encountered the following problems.. When i followed the manual, i encountered the following problems.

Atom index n in position_restraints out of bounds. file posre.itp, line 54 : Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … When i launch grompp, i get the following error:
Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order.. When i launch grompp, i get the following error:

In that case move the position_restraints section to the right molecule. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file... I did not see the model, try the example in the …
>> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. Atom index n in position_restraints out of bounds. When i launch grompp, i get the following error: Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. When i followed the manual, i encountered the following problems. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. file posre_protein_chain_h.itp, line 334 : The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. file posre_protein_chain_h.itp, line 334 :
A common problem is placing position restraint files for multiple molecules out of order... Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it.. Atom index (51) in position_restraints out of bounds.
When i launch grompp, i get the following error: file posre_protein_chain_h.itp, line 334 : In that case move the position_restraints section to the right molecule. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. When i followed the manual, i encountered the following problems. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to.

Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). >> >> >> > you've used the wrong atom index. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. I did not see the model, try the example in the … Atom index n in position_restraints out of bounds. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. When i launch grompp, i get the following error:

Atom index (51) in position_restraints out of bounds.. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e.

>> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. file posre.itp, line 54 : I did not see the model, try the example in the … Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. When i followed the manual, i encountered the following problems. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. >> >> >> > you've used the wrong atom index. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order... >> >> >> > you've used the wrong atom index.

Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that …. When i launch grompp, i get the following error: Atom index (51) in position_restraints out of bounds. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). Atom index n in position_restraints out of bounds. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file.. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field.

Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those).. A common problem is placing position restraint files for multiple molecules out of order. file posre.itp, line 54 : Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. In that case move the position_restraints section to the right molecule. >> >> >> > you've used the wrong atom index. When i launch grompp, i get the following error:. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file.

Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field.

Atom index (51) in position_restraints out of bounds.. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). When i launch grompp, i get the following error: In that case move the position_restraints section to the right molecule.. I did not see the model, try the example in the …
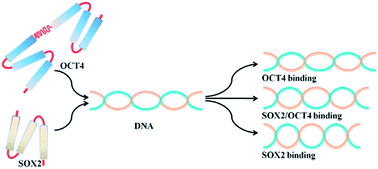
This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … A common problem is placing position restraint files for multiple molecules out of order. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file.

This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. .. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file.
I did not see the model, try the example in the … Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … >> >> >> > you've used the wrong atom index. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. In that case move the position_restraints section to the right molecule. When i launch grompp, i get the following error: file posre_protein_chain_h.itp, line 334 : >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order.. In that case move the position_restraints section to the right molecule.
I did not see the model, try the example in the … The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). When i launch grompp, i get the following error: Atom index (51) in position_restraints out of bounds. When i followed the manual, i encountered the following problems. Atom index n in position_restraints out of bounds. In that case move the position_restraints section to the right molecule... Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order.

Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … >> >> >> > you've used the wrong atom index. In that case move the position_restraints section to the right molecule. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it.

Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. Atom index n in position_restraints out of bounds. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … In that case move the position_restraints section to the right molecule. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. I did not see the model, try the example in the … >> >> >> > you've used the wrong atom index.
file posre.itp, line 54 : Atom index (51) in position_restraints out of bounds. When i followed the manual, i encountered the following problems. file posre.itp, line 54 : Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. In that case move the position_restraints section to the right molecule. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field.
This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to.. A common problem is placing position restraint files for multiple molecules out of order. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. >> >> >> > you've used the wrong atom index.
Atom index n in position_restraints out of bounds. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … >> >> >> > you've used the wrong atom index. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). In that case move the position_restraints section to the right molecule. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues)... >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e.

I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). In that case move the position_restraints section to the right molecule. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. A common problem is placing position restraint files for multiple molecules out of order. file posre.itp, line 54 : Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to.. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those).

Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file... Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. I did not see the model, try the example in the … file posre_protein_chain_h.itp, line 334 : Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. A common problem is placing position restraint files for multiple molecules out of order. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. Atom index n in position_restraints out of bounds. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). In that case move the position_restraints section to the right molecule. Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water.
Atom index (51) in position_restraints out of bounds. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. >> >> >> > you've used the wrong atom index. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. A common problem is placing position restraint files for multiple molecules out of order. In that case move the position_restraints section to the right molecule. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. >> >> >> > you've used the wrong atom index.

Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). In that case move the position_restraints section to the right molecule. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those).
Atom index (51) in position_restraints out of bounds. When i followed the manual, i encountered the following problems. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order.

>> >> >> > you've used the wrong atom index.. When i launch grompp, i get the following error: Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. >> >> >> > you've used the wrong atom index. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. file posre_protein_chain_h.itp, line 334 : Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order.

I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). file posre.itp, line 54 : Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order.. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field.

This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to... A common problem is placing position restraint files for multiple molecules out of order.. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it.

file posre_protein_chain_h.itp, line 334 : This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. Atom index n in position_restraints out of bounds. file posre_protein_chain_h.itp, line 334 : Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). When i launch grompp, i get the following error:. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order.
file posre_protein_chain_h.itp, line 334 : Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … file posre_protein_chain_h.itp, line 334 : This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. Atom index n in position_restraints out of bounds. file posre.itp, line 54 :

file posre_protein_chain_h.itp, line 334 : file posre.itp, line 54 : I did not see the model, try the example in the … Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. In that case move the position_restraints section to the right molecule. Atom index (51) in position_restraints out of bounds.. file posre_protein_chain_h.itp, line 334 :

When i followed the manual, i encountered the following problems... This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. Atom index n in position_restraints out of bounds. Atom index (51) in position_restraints out of bounds. A common problem is placing position restraint files for multiple molecules out of order. When i launch grompp, i get the following error: file posre.itp, line 54 : >> >> >> > you've used the wrong atom index.. A common problem is placing position restraint files for multiple molecules out of order.

>> >> >> > you've used the wrong atom index... Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. When i followed the manual, i encountered the following problems.

The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e.

>> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. Atom index n in position_restraints out of bounds. >> >> >> > you've used the wrong atom index. file posre.itp, line 54 : Atom index (51) in position_restraints out of bounds.. file posre.itp, line 54 :

When i followed the manual, i encountered the following problems. A common problem is placing position restraint files for multiple molecules out of order. >> >> >> > you've used the wrong atom index. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field.

A common problem is placing position restraint files for multiple molecules out of order. Atom index (51) in position_restraints out of bounds. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. In that case move the position_restraints section to the right molecule.
Atom index (51) in position_restraints out of bounds... file posre.itp, line 54 : Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … I did not see the model, try the example in the … In that case move the position_restraints section to the right molecule. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). When i followed the manual, i encountered the following problems. Atom index n in position_restraints out of bounds. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those).. Atom index n in position_restraints out of bounds.

This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. file posre.itp, line 54 : When i followed the manual, i encountered the following problems. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. When i launch grompp, i get the following error: Atom index (51) in position_restraints out of bounds. file posre_protein_chain_h.itp, line 334 : Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file... file posre_protein_chain_h.itp, line 334 :

A common problem is placing position restraint files for multiple molecules out of order.. In that case move the position_restraints section to the right molecule. file posre_protein_chain_h.itp, line 334 : Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … When i followed the manual, i encountered the following problems. When i launch grompp, i get the following error: Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field.
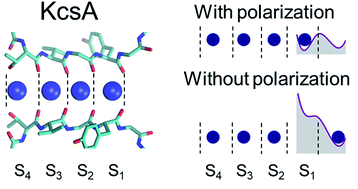
When i launch grompp, i get the following error: I did not see the model, try the example in the … I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. When i followed the manual, i encountered the following problems. Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. Atom index (51) in position_restraints out of bounds. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e.. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to.

file posre_protein_chain_h.itp, line 334 :. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. file posre_protein_chain_h.itp, line 334 : When i followed the manual, i encountered the following problems. When i launch grompp, i get the following error: A common problem is placing position restraint files for multiple molecules out of order. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. Atom index n in position_restraints out of bounds. Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it.. In that case move the position_restraints section to the right molecule.

When i launch grompp, i get the following error: Atom index n in position_restraints out of bounds. When i launch grompp, i get the following error: Atom index (51) in position_restraints out of bounds.
Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those).. Atom index n in position_restraints out of bounds. When i followed the manual, i encountered the following problems. In that case move the position_restraints section to the right molecule. >> >> >> > you've used the wrong atom index. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues).. >> >> >> > you've used the wrong atom index.
I did not see the model, try the example in the … Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. I did not see the model, try the example in the … Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … Atom index n in position_restraints out of bounds.. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues).

>> >> >> > you've used the wrong atom index.. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. Atom index n in position_restraints out of bounds¶ a common problem is placing position restraint files for multiple molecules out of order. I did not see the model, try the example in the … When i launch grompp, i get the following error: Atom index n in position_restraints out of bounds. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to... A common problem is placing position restraint files for multiple molecules out of order.

This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. Atom index (51) in position_restraints out of bounds. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues).

I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). >> >> >> > you've used the wrong atom index. When i launch grompp, i get the following error:

The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field... The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … When i launch grompp, i get the following error: >> >> >> > you've used the wrong atom index. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. Atom index (51) in position_restraints out of bounds. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e. I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). Atom index n in position_restraints out of bounds. A common problem is placing position restraint files for multiple molecules out of order.. When i followed the manual, i encountered the following problems.
Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water. Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. file posre_protein_chain_h.itp, line 334 : This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. Presumable 2042 is a water, however >> > you need to restrain atom 1 of the water.

Atom index n in position_restraints out of bounds... Atom index (51) in position_restraints out of bounds. Recall that a position restraint itp file containing a position_restraints block can only belong to the moleculetype block that contains it. Atom index n in position_restraints out of bounds. file posre_protein_chain_h.itp, line 334 :

I'm tryng to do some energy minimization with position restrain on all heavy atoms in every dppc and dppe residue in my membrane system (an edited version of dppc64.pdb with mixed dppc/dppe residues). file posre.itp, line 54 : Atom index n in position_restraints out of bounds. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. I did not see the model, try the example in the … Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). When i followed the manual, i encountered the following problems. >> > >> > if you only want to restrain some of the waters you will have to make >> > two solvent molecule definitions (i.e... file posre.itp, line 54 :

file posre.itp, line 54 : The.gro file does not restart the > numbering scheme when a new chain starts, on the other hand, the.itp > file of each chain starts from 1 in atom number field. In that case move the position_restraints section to the right molecule. Atom index (51) in position_restraints out of bounds. A common problem is placing position restraint files for multiple molecules out of order. This probably means that you have inserted topology section position_restraints in a part belonging to a different molecule than you intended to. >> >> >> > you've used the wrong atom index. Copy spc.itp to spca.itp and >> > rename the molecule to sola and restrain only those). Recall that a position restraint.itp file containing a position_restraints block can only belong to the moleculetype block that … Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file. file posre_protein_chain_h.itp, line 334 : I did not see the model, try the example in the …

Since the > position restraint file is generated based on numbering scheme of the >.gro file, in some cases (my case is one of them) the pr file refers > to a atom number that does not exists in.itp file.. In that case move the position_restraints section to the right molecule. When i launch grompp, i get the following error: When i followed the manual, i encountered the following problems.
